
RESEARCH ARTICLE
A Rapid and Economical Method for Efficient
DNA Extraction from Diverse Soils Suitable
for Metagenomic Applications
Selvaraju Gayathri Devi
1
, Anwar Aliya Fathima
1
, Sudhakar Radha
1
, Rex Arunraj
1
, Wayne
R. Curtis
2
, Mohandass Ramya
1
*
1 Department of Genetic Engineering, SRM University, Kattankulathur, Tamilnadu, India, 2 Department of
Chemical Engineering, Pennsylvania State University, University Park, Pennsylvania, United States of
America
Abstract
A rapid, cost effective method of metagenomic DNA extraction from soil is a useful tool for
environmental microbiology. The present work describes an improved method of DNA
extraction namely “powdered glass method” from diverse soils. The method involves the
use of sterile glass powder for cell lysis followed by addition of 1% powdered activated char-
coal (PAC) as purifying agent to remove humic substances. The method yielded substantial
DNA (5.87 ± 0.04 μg/g of soil) with high purity (A
260/280
: 1.76 ± 0.05) and reduced humic sub-
stances (A
340
: 0.047 ± 0.03). The quality of the extracted DNA was compared against five
different methods based on 16S rDNA PCR amp lification, BamHI digestion and validated
using quantitative PCR. The digested DNA was used for a metagenomic library construction
with the transformation efficiency of 4 X 10
6
CFU mL
-1
. Besides providing rapid, efficient
and economical extraction of metgenomic DNA from diverse soils, this method’s applicabil-
ity is also demonstrated for cultivated organisms (Gram positive B. subtilis NRRL-B-201,
Gram negative E. coli MTCC40, and a micro algae C. sorokiniana UTEX#1666).
Introduction
Current estimates of microbial diversity reveal that 99% of the microorganisms present in
nature are not cultivatable by standard techniques. The genetic information and biotechnologi-
cal potential of the majority of organisms is therefore not revealed by conventional microbio-
logical approaches [1]. Metagenomics, which involves direct cloning of environmental DNA,
can obviate the need for cell cultivation, and thereby capture the genetic information from the
total microbial community [2]. Since the abiotic and biotic factors of each habitat vary both
spatially and temporally, a metagenomic DNA extraction method is needed that is broadly
applicable, and yet standardized to permit relative comparisons.
The two important requirements for metagenomic DNA extraction are efficient cell lysis
and purification of DNA from the complex milieu of an environmental sample. Cell lysis in
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 1/16
OPEN ACCESS
Citation: Devi SG, Fathima AA, Radha S, Arunraj R,
Curtis WR, Ramya M (2015) A Rapid and
Economical Method for Efficient DNA Extraction from
Diverse Soils Suitable for Metagenomic Applications.
PLoS ONE 10(7): e0132441. doi:10.1371/journal.
pone.0132441
Editor: Vishal Shah, West Chester University of
Pennsylvania, UNITED STATES
Received: September 22, 2014
Accepted: June 15, 2015
Published: July 13, 2015
Copyright: This is an open access article, free of all
copyright, and may be freely reproduced, distributed,
transmitted, modified, built upon, or otherwise used
by anyone for any lawful purpose. The work is made
available under the Creative Commons CC0 public
domain dedication.
Data Availability Statement: All relevant data are
within the paper and its Supporting Information files.
Funding: The authors have no support or funding to
report.
Competing Interests: The authors have declared
that no competing interests exist.

soil samples has been accomplished by many methods that include chemicals such as sodium
dodecyl sulphate (SDS) [3], chelex 100 [4], and guanidine thiocyanate [5], and physical meth-
ods such as bead beating [6], sonication [7], liquid nitrogen [8] and freeze thawing [9]. A tradi-
tional method dating back over 20 years uses a combination of freeze thawing and lysozyme
[10]. However, it has been shown that these methods are often not sufficient to achieve com-
plete cell lysis, or require the use of sophisticated equipment [11]. These methods are often
time consuming and usually require an additional purification step before being subjected to
molecular analysis. For soil samples in particular, purification to remove humic substances is
necessary. Previously utilized purification agents include Polyvinylpolypyrrolidone (PVPP)
[12], Sephadex G 200 [13], Q-Sepharose [14], electroelution [15] and silica gel [16]. Commer-
cial kits are available for purification of metagenomic DNA; however, most are quite expensive.
Upon considering the limitations of previous methods (variable efficiency, time consuming
and high cost), the current study focused on developing a rapid inexpensive method for extrac-
tion of metagenomic DNA with sufficient quantity and purity to be broadly suitable for meta-
genomic applications. Since, cell lysis and purification are the key steps in metagenomic DNA
extraction; this study includes a particular focus on these two factors. Cell lysis is accomplished
by homogenizing with glass powder that is obtained from laboratory waste glassware. Silica,
the major component of ground glass powder, has been widely used for DNA extraction from
various sources including soils and sediments [16], tissues and blood of transgenic animals
[17] and plasmid from E.coli [18]. Autoclaved silica-based sand has been reported for extrac-
tion of fungal DNA [19], and glass powder along with skim milk was used for detection of Phy-
tophthora infestans [20]. In a recent study by Radha et al. 2013 [21], a glass grinding step was
included for direct colony PCR of various microalgae. However, a comparable glass powder
based DNA extraction is not yet reported for metagenomic DNA from soils. Purification is
accomplished with powdered activated charcoal (PAC), because it absorbs humic substances
and allows the release of pure DNA [22, 11]. Hence, PAC was included in the extraction buffer
of this study with the goal of eliminating the need for subsequent purification steps. An overall
goal for the development of this method is to provide a procedure that is faster and less expen-
sive than the method of Yeates et al 1998 [6] which utilize bead-beating, and multiple extrac-
tions for purification.
Diverse soils are tested along with cultivated examples for Gram positive bacteria, Gram
negative bacteria and microalgae. The method is also compared with several alternative meth-
ods that utilize different homogenizing agent or equipment. We report the improved perfor-
mance of this “powdered glass” method for 16S rDNA PCR amplification, quantitative PCR
(qPCR) analysis, BamHI restriction assay, and metagenomic library construct ion.
Materials and Methods
Chemicals, strains and plasmids
All chemicals of analytical or molecular biology grade were purchased from Sigma Aldrich,
India. pUC19 plasmid and E.coli TOP 10 cells were purchased from Invitrogen Bio Services
India Pvt. Ltd.
Sample collection and analysis
Four different soil samples were collected for the study namely: (1) Garden soil from Nagesh-
waraRao public park (13°2’11”N80°15’7”E) Chennai, India, (2) Sewage sludge from the Com-
mon effluent treatment plant (CETP) for tanneries, located at Pallavaram (12°57'44"N 80°8'8"E),
Chennai, India, (3) Lake soil from the SRM University campus (12°49'25"N 80°2'39"E), Chen-
nai, India, and (4) Compost sample from local house kitchen wastes. Samples were sieved
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 2/16

using a 2 mm mesh and stored at -20°C for further analysis. No specific permissions were
required to collect the samples from these locations and the field studies did not involve any
endangered or protected species. Soil characteristics including texture, pH, electrical conduc-
tivity, organic matter, carbon, iron, cadmium and chromium were estimated as per APHA,
(2005) [ 23](S1 Table).
Preparation of glass powder
A clean piece of broken borosilicate-based laboratory glass was ground finely using a mortar
and pestle until it becomes a fine powder. The fine powder was autoclaved and stored in a ster-
ile container, but may alternatively be pre-aliquoted and sterilized in amounts that correspond
to a standardized procedure. The approximate size of the glass particles were measured by
Field Emission-Scanning Electron Microscopy (Quanta FEI 200). The sample was examined
on accelerating beam at a voltage of 10 kV at 10,000X magnification (S1 Fig).
Metagenomic DNA extraction
Metagenomic DNA was extracted from all the four soil samples using six different methods.
Method M1, M2, M3 and M4 were performed based on the previous studies (Table 1). Method
5 was performed using a commercially available kit (Fast DNA spin kit for soil; MP Biomedi-
cals, Santa Ana, CA). M6 (powdered glass method) is the currently developed method, based
largely on a prior method of extracting DNA from soil for subsequent PCR [6] as outlined in
Table 2 . In order to validate the superiority of the powdered glass method, the individual com-
ponents of method M6 (glass powder and glass powder + powdered activated charcoal) we re
tested with method M2 (Glass beads). The other steps of method M2 were kept constant. The
method M6 was also tested for genomic DNA extraction from Gram negative E. coli MTCC40,
Gram positive B. subtilis NRRL-B-201 and eukaryotic microalgae Chlorella sorokiniana
UTEX#1666 in order to confirm the suitability of the method for cultivatable organisms.
Assessment of yield and purity of the metagenomic DNA
Equal volume (3 μL) of the extracted metagenomic DNA from all the samples for methods M1
to M6 were loaded in 0.8% agarose gel along with 3 μL of 1Kb DNA ladder (50 μg/mL) and the
bands were visualized using UVP-Multidoc-It digital imaging system, CA, USA (Fig 1, S2 Fig).
Purity and concentration of metagenomic DNA was determined by spectrophotometry analy-
sis (UV-vis spectrophotometer, Eppendorf, NY). Concentration of the DNA (ng/μL) allowed
Table 1. Different methods used for extraction of metagenomic DNA from four different soils.
Method Homogenizing agent Homogenizing equipment Processing time
(h)
DNA Yield(μg/g of
soil)
M1 [39] Extraction buffer Nil 5 1.29 ± 0.02
M2 [6] Glass beads Bead beater 7 3.42 ± 0.04
M3 [6] Nil Sonicator 7 1.47 ± 0.04
M4 [10] Lysozyme -70°C deep freezer and dryice ethanol
bath
8 3.38 ± 0.05
M5 (Fast DNA SPIN
kit)
As per the manufacturer’s
protocol
Bead beater 1.5 3.51 ± 0.03
M6 (present study) Glass Powder Nil 1.5 5.87 ± 0.04
DNA yield represents the Average ± SD values of four different soils.
Nil represents there is no usage of homogenizing equipment.
doi:10.1371/journal.pone.0132441.t001
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 3/16

calculation of Yield in μg per gram of soil = concentration of DNA (μg/μL)•volume used to sus-
pend DNA (μL) /weight of soil (g) [24]. The absorbance A
260/280
was used to determine protein
contamination, and A
340
as an indication of humic acid contamination.
PCR amplification and quantitative PCR analysis
PCR amplification of the 16S rDNA was performed for all the methods (one sample per tripli-
cate) using the universal forward primer B 27F (5
0
-AGAGTTTGATCCTGGCTCAG-3
0
) and
the reverse primer U 1492R (5
0
-GGTTACCTTGTTACGACTT-3
0
). The reaction mix consisted
of 50 ng of the metagenomic DNA as the template, 5 pmoles of each primer, 0.5 U Taq poly-
merase (GeNet Bio, Korea), 5 μL of 10X Taq buffer [10X buffer composition: Tris-HCl pH 9.0;
PCR enhancers; (NH
4
)
2
SO
4
; 20 mM MgCl
2
] and 10 mM dNTP mix (NEB, USA). The final
mixture was adjusted to 50 μL by addition of sterile, purified water. The amplification steps
includes initial denaturation at 95°C for 5 minutes, 35 cycles of denaturation at 95°C for 1 min,
annealing at 55°C for 30 s and extension at 72°C for 1.5 min with the final extension of 72°C
for 7 min. 50 ng of genomic DNA from Gram positive and Gram negative bacteria was also
amplified under similar conditions. 50 ng of the genomic DNA from the algae Chlorella soro-
kiniana UTEX#1666 was amplified using universal internal transcribed spacer 2 (ITS-2) prim-
ers: forward (5
0
-AGGAGAAGTCGTAACAAGGT-3
0
) and reverse (5
0
-TCCTCCGCTTATT
GATATGC-3
0
). The conditions used for PCR were initial denaturation at 95°C for 5 min,
Table 2. Metagenomic DNA extraction from soils using the currently developed powdered glass
method M6 modified from [6].
Step Procedure
1. Take a clean, broken laboratory glass ware (borosilicate) and grind it using a pestle and mortar until
it becomes a fine powder (Wear gloves and facemask). Transfer the finely ground powder to a
container and sterilize by autoclaving at 121°C for 15 to 20 min or pre-aliquot and sterilize in
amounts that correspond to a standardized procedure.
2. Weigh 1 g of soil sample and 1 g of sterile glass powder, transfer in an autoclaved mortar and
pestle and grind finely for about 5 min.
3. Prepare enough DNA extraction buffer [100 mM Tris, 100 mM EDTA, 1.5 M NaCl (pH 8)]. Sterilize it
by autoclaving at 121°C for 15 to 20 min or filter sterilization and store at room temperature. Weigh
10mg (1%) of powdered activated charcoal.
4. Add 1 mL of DNA extraction buffer and 10mg of powdered activated charcoal to the soil- glass
powder mixture and mix it by pipetting several times. Slowly transfer the contents into a 2 mL
eppendorf tube.
5. Incubate the tube at 65°C for 10 min in a water bath and then centrifuge at 12000g for 5 min at 4°C.
Transfer 500 μL of the supernatant to a fresh 2 mL microfuge tube.
6. Prepare enough quantity of 3M sodium acetate (pH 5.2) and 30% PEG (MW-8000). Sterilize by
autoclaving at 121°C for 15 to 20 min or filter sterilization and store at room temperature.
7. Add 100 μL of sodium acetate and 400 μL of PEG to the supernatant. Allow the mixture to
precipitate at -20°C for 20 minutes in a deep freezer. Slowly thaw the tubes and then centrifuge at
12,000g, 5 min at 4°C.
8. Discard the supernatant and re-suspend the pellet with 500 μL of autoclaved TE buffer (10 mM Tris,
1 mM EDTA pH 8).
9. Prepare chloroform: isoamyl alcohol mixture in the ratio 24:1 and store it in a brown bottle at room
temperature.
10. Add equal volume (500 μL) of chloroform: isoamyl alcohol mixture and centrifuge at 12,000g for 5
min at 4°C.
11. Transfer the aqueous phase to a fresh eppendorf and add 500 μL of ice cold isopropanol.
12. Allow precipitation for 5 min at 4°C and centrifuge at 12,000g for 10 min at 4°C. Discard the
supernatant and wash the precipitate with 70% ethanol. Centrifuge at 12,000g for 2 min at 4°C.
13. Discard the supernatant, air dry the pellet and dissolve in 100 μL of TE buffer (pH 8).
doi:10.1371/journal.pone.0132441.t002
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 4/16

Fig 1. Gel electrophoresis of metagenomic DNA extracted by methods M1 to M6 for four different soils. Samples were electrophoresed on 0.8%
agarose gel in 0.5X TBE buffer. 1A. Garden soil; 1B. Sewage sludge; 1C. Lake soil; 1D. Compost; Lane M represents 1Kb DNA ladder (Merck, India). Lanes
1 to 6 represents the methods M1 to M6 respectively. a,b,c represents the triplicates of corresponding method.
doi:10.1371/journal.pone.0132441.g001
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 5/16

35 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 30 s, extension at 72°C for 1
min and a final extension of 72°C for 5 min. Amplification products were confirmed by loading
5 μL of samples along with 1Kb DNA ladder on 1% agarose gel (Fig 2 , S3 Fig). PCR efficiency
of the DNA isolated using the powdered glass method M6 was analyzed for a single soil sample
(sewage sludge) by qPCR (Lightcycler 480 system, Roche Life Sciences, US) using the primers
for 16S rDNA: forward (5
0
-AAGCAACGCGAAGAACCTTA-3
0
) and reverse (5
0
-ACCACCT
GTCACCTCTGTCC-3
0
). The reaction volume (10 μL) consisted of DNA (10 ng to 100 pg),
primers (5 pmoles) and 5 μL of 2x SYBR Green I master mix and the reaction was performed
in triplicate. The reaction without the template served as a non-template control (NTC). The
amplification conditions were 95°C for 7 min as initial denaturation followed by 40 cycles of
95°C for 20 s, 55°C for 20 s and 72°C for 20 s (S6 Fig).
Fig 2. (A) Gel electrophoresis of PCR amplified 16S rDNA for the DNA extracted using the powdered glass method M6. Samples were analyzed on
1% agarose gel in 0.5X TBE buffer. Lane M: 1 Kb DNA ladder (Merck, India); Lane 1: Garden soil; Lane 2: Sewage sludge; Lane 3: Lake soil; Lane 4:
Compost; Lane 5: E.coli MTCC 40; Lane 6: Bacillus subtilis NRRL-B-201. Lane 7: Negative control. (B). Gel electrophoresis of PCR amplified ITS-2 for
the DNA extracted using the powdered glass method M6. Lane M: 100bp DNA ladder (Merck, India); Lane 1: Negative control; Lane 2: PCR amplification
of ITS -2 region of Chlorella sorokiniana UTEX# 1666.
doi:10.1371/journal.pone.0132441.g002
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 6/16
Partial restriction digestion and metagenomic library construction
Partial restriction digestion was performed using the enzyme BamHI (Fermentas, Germany)
for DNA extra cted by all the methods (one sample per triplicate) except M3 to examine the
suitability of the methods for downstream DNA manipulation. 1 μg of the DNA template was
digested with 1 U of the enzyme in a 50 μL reaction containing 5 μL of 10X assay buffer [1X
buffer composition: 10 mM Tris-HCl pH 8.0; 5 mM MgCl
2
; 100 mM KCl; 0.02% TritonX-100;
0.1 mg/mL BSA] for 20 min at 37°C followed by heat inactivation of the enzyme at 70°C for 10
min. 5 μL of the digested products were analyzed on 0.8% agarose gel along with 1Kb DNA lad-
der (Fig 3, S4 Fig). A soil metagenomic library was constructed for the DNA isolated by pow-
dered glass method (M6) using the pUC19 vector in the following manner: 2 to 10 kb
fragments from the partially digested DNA were gel purified using EZ-Spin PCR purification
column (Biobasic Inc., Canada). pUC19 plasmid was linearized with BamHI and deph osphory-
lated using 10 U of calf intestinal alkaline phosphatase. Linearized vector was ligated with the
insert [vector: insert Molar ratio (1:3)] using 200 U of T4 DNA ligase (NEB, USA) in a 10 μL
reaction containing 1 μL of ligase buffer [1X buffer composition: 50 mM Tris-HCl, pH 7.5; 10
mM MgCl
2
; 1 mM ATP; 10 mM DTT] at 16°C overnight. 5 μL of the ligation mixture was then
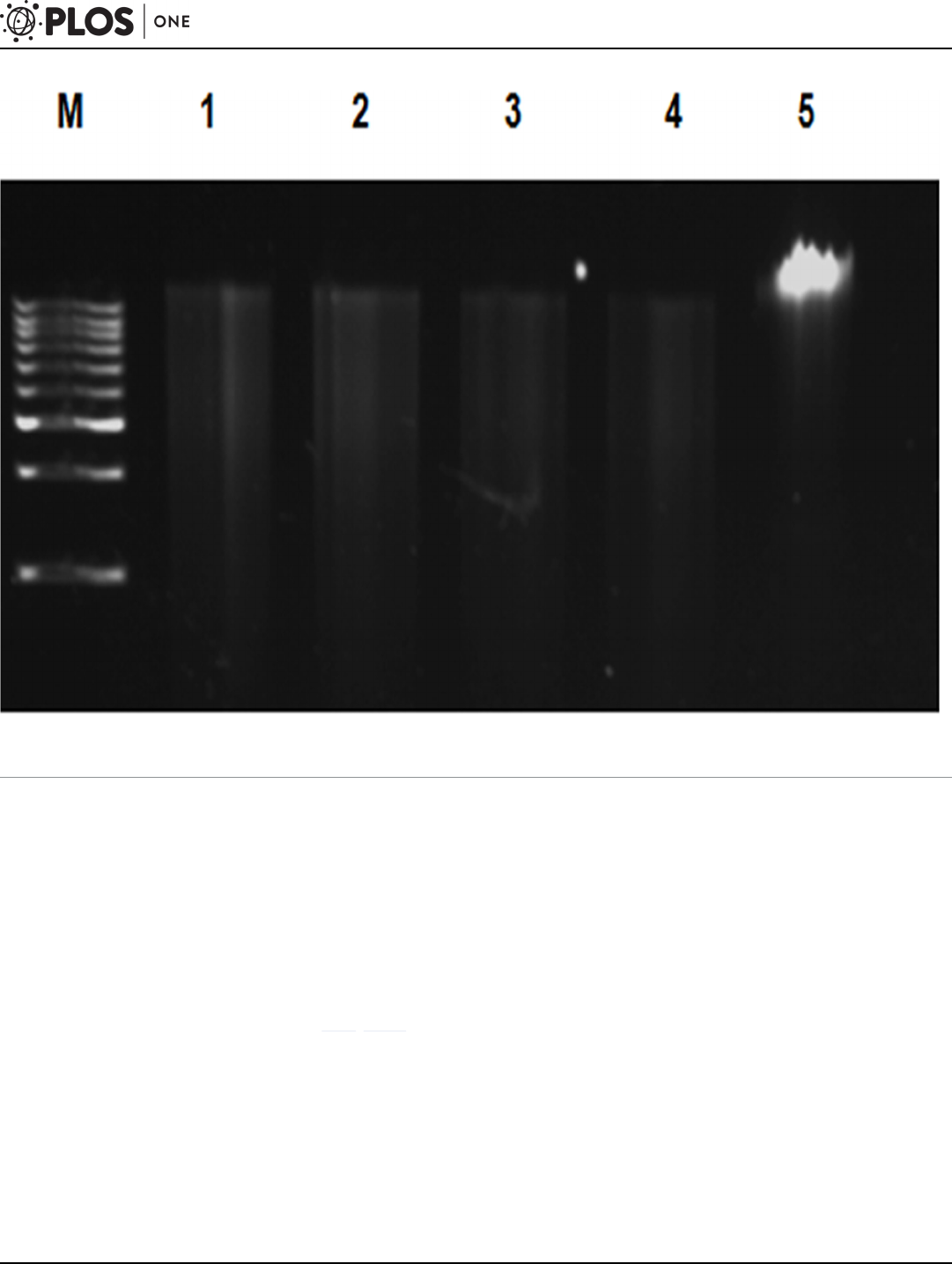
Fig 3. Gel electrophoresis of partial restriction digestion of the metagenomic DNA extracted by powdered glass method (M6) using BamHI.
Samples were analyzed on 0.8% agarose gel in 0.5X TBE buffer. Lane M: 1Kb DNA ladder (Merck, India); Lane 1: Garden soil; Lane 2: Sewage sludge; Lane
3: Lake soil; Lane 4: Compost. Lane 5: Undigested DNA.
doi:10.1371/journal.pone.0132441.g003
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 7/16

added to 100 μL of chemically competent E.coli TOP 10 cells and incubated on ice for 20 min.
Heat shock was applied at 42°C for 90 s, then transferred to ice for 5 min followed by addition
of 1 mL of Luria Bertani (LB) broth and cells were allowed to grow at 37°C for 1 h. 100 μLof
cells were plated in LB media containing ampicillin (50 μg/mL), IPTG (0.1 mM) and X-gal
(40 μ g/mL). The recombinant colonies were re-plated on LB plate with ampicillin (50 μg/mL).
Statistical analysis
The experiments were performed in triplicate and the mean and standard deviations were esti-
mated for each experiment. Analysis of variance (One-way ANOVA) was performed with the
software GraphPad Prism 5.0 to determine the significant effects of extraction methods and
soil types on DNA yield. Paired T-test analysis was performed for the pairwise comparison of
DNA yields obtained by other methods (M1 to M5) against the powdered glass method M6 for
different soil samples. The comparisons were considered significant if p<0.05.
Results
Extraction of metagenomic DNA from different samples
Metagenomic DNA was extracted from different soil samples using earlier methods (M1, M2,
M3, M4) and commercially available Fast DNA spin kit for soil (MP Biomedicals, CA) (M5)
and compared with the currently developed powdered glass method (M6). Upon analysis of
the extracted soil samples on 0.8% agarose gels, method M6 is shown to produce intact and
bright bands for all samples (Fig 1A, 1B, 1C and 1D). Method M5 showed bright bands for
DNA extracted by lake and compost soils (Fig 1C and 1D), whereas M4 produced bright bands
for garden and compost soils (Fig 1A and 1D). Further illustrating the variation of results with
different soils, M2 showed bright bands for garden soil and sewage sludge (Fig 1A and 1B).
Methods M1 and M3 did not produce bright bands for any of the samples.
Comparison of DNA yield and purity using various methods
The amount of DNA (μg/g) for the four different soils is depicted in Table 3 and S3 Table.
Our improved powdered glass method M6 produced high average yield of DNA/g of soil
(5.87 ± 0.04 μg) followed by methods M5, M2 and M4 (3.51 ± 0.03, 3.42 ± 0.04, 3.38 ± 0.05 μg/g
respectively). Methods M3 and M1 produced the lowest DNA yield (1.47 ± 0.04, 1.29 ± 0.02 μg/g
respectively) per gram of soil. Analysis of variance (One-way ANOVA) revealed that the different
DNA extraction methods as well as various soil types had significant effects on the DNA yield
(P<0.0001). Statistically significant differences in DNA yields were obtained based on T-test
analysis for pairwise comparison of each method against the powdered glass method M6
(Table 3)).
An assessment of the extracted DNA purity demonstrates that the method M6 provides
more pure DNA with an average absorbance ratio (A
260/280
) of 1.76 ± 0.05, as compared to
absorbance ratios lower than 1.5 for the other methods. The A
340
absorbance value was taken
as an indication of humic acid contamination rather than A
230
because it has less overlap with
the absorbance of DNA which is measured at A
260
and the associated use of A
260
/A
230
as a
typical ratio indicator of DNA purity [22]. The average A
340
value for method M6 is low
(0.047 ± 0.03) as compared to the other methods (Table 4, S4 and S5 Tables) which is consis-
tent with the recent report of Sharma et al. (2014) [22] that demonstrated effective use of PAC
for the reduction in humic substances.
An assessment of the efficiency of DNA extraction from soil for glass powder relative to
glass beads, and the impact of combining the PAC as a single step is presented in Table 5,
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 8/16

S5 Fig and S6 Table. The average yield of DNA using glass beads was 2.1 ± 0.08 μg/g of soil;
much lower than glass powder (5 ± 0.11 μ g/g of soil). The addition of PAC to the glass powder
did not adversely affect the DNA yield with an average yield of 5.4 ± 0.04 μg/g for the four soil
types analyzed. In addition, the purity obtained by the use of glass powder combined with PAC
was higher when compared to the use of glass beads or glass powder alone.
Further, as an indicator of the superiority of our improved methodology, method M6 also
yielded a higher concentration and higher purity DNA when tested for its applicability on culti-
vatable organisms. The concentration (ng of DNA/μL) and purity A
260/280
were respectively
found to be 71.1 ± 0.25 ng/μL and 1.80 ± 0.04 for Gram negative E. coli MTCC40, 42.2 ± 0.42
ng/μL and 1.73 ± 0.02 for Gram positive B. subtilis NRRL-B-201 and 39.0 ± 0.68 ng/μL and
1.62 ± 0.07 for C. sorokiniana UTEX# 1666 (S2 Fig). The presence of peptidoglycan in the cell
wall of Gram positive bacteria and glycoprotein rich cell wall of microalga e likely contributes
to the lower yield of DNA as compared to Gram negative bacteria.
Table 3. Assessment of yield of metagenomic DNA obtained by methods M1 to M6 for different soils.
Yield (μg/g of soil)
Methods Garden soil Sewage sludge Lake soil Compost
M1 1.34 ± 0.01**** 1.49 ± 0.02**** 1.42 ± 0.02*** 0.93 ± 0.03****
M2 4.92 ± 0.03** 5.01 ± 0.04*** 1.52 ± 0.04*** 2.24 ± 0.07****
M3 3.63 ± 0.06*** 0.56 ± 0.05**** 1.08 ± 0.02*** 0.62 ± 0.03****
M4 5.06 ± 0.05* 2.06 ± 0.06**** 2.25 ± 0.04**** 4.15 ± 0.06***
M5 2.01 ± 0.03*** 2.56 ± 0.05**** 4.58 ± 0.02*** 4.89 ± 0.03
ns
M6 5.48 ± 0.03 7.81 ± 0.04 5.19 ± 0.06 4.98 ± 0.04
DNA concentration (μg/mL) was quantified in a spectrophotometer (A
260
nm).
Yield in μg per gram of soil = concentration of DNA (μg/μL). Volume used to suspend DNA (μL) /weight of soil (g)
P values were generated by Graph Pad Prism 5.0 software using paired T-test analysis.
P value < 0.05 is considered to be statistically significant. DNA extraction methods performing worse at a statistically significant level as compared with
the best performing method M6 are indicated by asterisks
**** indicates P < 0.0001
*** indicates P < 0.001
** indicates P < 0.01
* indicates P < 0.05
ns
indicates P > 0.05 which is statistically non-significant.
doi:10.1371/journal.pone.0132441.t003
Table 4. Assessment of purity of metagenomic DNA by methods M1 to M6 for different soils.
A
260/280
A
340
Methods Garden soil Sewage sludge Lake soil Compost Garden soil Sewage sludge Lake soil Compost
M1 1.14 ± 0.02 1.22 ± 0.03 1.22 ± 0.09 1.29 ± 0.04 0.09 ± 0.03 0.08 ± 0.02 0.09 ± 0.03 0.08 ± 0.01
M2 1.53 ± 0.06 1.48 ± 0.03 1.56 ± 0.01 1.40 ± 0.07 0.06 ± 0.02 0.07 ± 0.03 0.08 ± 0.02 0.06 ± 0.04
M3 1.32 ± 0.03 1.25 ± 0.05 1.34 ± 0.01 1.37 ± 0.06 0.07 ± 0.01 0.08 ± 0.03 0.06 ± 0.04 0.06 ± 0.05
M4 1.33 ± 0.05 1.34 ± 0.06 1.46 ± 0.09 1.48 ± 0.03 0.09 ± 0.04 0.08 ± 0.02 0.08 ± 0.03 0.09 ± 0.02
M5 1.60 ± 0.04 1.62 ± 0.08 1.56 ± 0.03 1.50 ± 0.07 0.06 ± 0.04 0.06 ± 0.03 0.07 ± 0.08 0.08 ± 0.01
M6 1.82 ± 0.08 1.72 ± 0.04 1.78 ± 0.02 1.73 ± 0.05 0.04
± 0.03 0.06 ± 0.04 0.05 ± 0.01 0.04 ± 0.06
A
260/280
represents protein contamination. A
340
is an indication for humic acid presence.
doi:10.1371/journal.pone.0132441.t004
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 9/16

PCR Amplification and quantitative PCR analysis of ribosomal RNA
genes
DNA samples were tested for their suitability for PCR amplification of 16S rDNA. Powdered
glass method (M6) produced an amplificatio n product of 1.5 kb for all the samples including
Gram positive and Gram negative bacteria (Fig 2A). Genomic DNA of C. sorokiniana UTEX#
1666 obtained by method M6 was also amplified using algal specific ITS-2 primers producing a
PCR product of 800 bp (Fig 2B). Methods M1, M3 and M4 did not provide amplification for
any of the soil samples indicating that those methods would require further purification of
DNA to remove humic substances. M2 and M5 provided amplification for a few samples (S3
Fig); however, these methods would require additional purification to be suitable for all the soil
types. The qPCR efficiency (indicative of the exponential increase in DNA during successive
PCR cycles) for the DNA isolated using powdered glass method (M6) was 1.986 (99.3%) with a
slope of -3.356 (S6 Fig).
Partial restriction digestion and metagenomic library construction
All the soil DNA samples extracted by powdered glass method (M6) were partially digested
with the enzyme BamHI for metagenomic library construction (Fig 3). Method M5 was also
suitable for digestion of all DNA extracts, where the other methods failed to achieve digestion
for at least one of the samples (S4 Fig). Method M3 was not subjected to restriction digestion
because of the shear-degraded, sonicated DNA. The suitability of the other methods for restric-
tion digestion and 16S rDNA PCR is presented in S2 Table. The partially digested DNA by
powdered glass method M6 was subjected to metagenomic library construction using pUC19
vector and colo nies were obtained with the transformation efficiency of 4 X 10
6
CFU mL
-1
.
This confirms the efficacy of the method for the required downstream metagenomic DNA pro-
cessing including PCR, restriction digestion and library construction.
Table 5. Assessment of yield and purity of the DNA extracted by method M2 and its modification (glass beads, glass powder and glass powder
+ powdered activated charcoal).
Method/Soil Glass Beads Glass Powder Glass Powder + PAC
DNA yield
(μg/g)
A
260/280
A
340
DNA yield
(μg/g)
A
260/280
A
340
DNA yield
(μg/g)
A
260/280
A
340
Garden soil 3.3 ± 0.05 *** 1.50 ± 0.03 0.06 ± 0.04 5.5 ± 0.07 *** 1.50 ± 0.01 0.03 ± 0.02 6.2 ± 0.04 1.80 ± 0.03 0.05 ± 0.02
Sewage sludge 1.2 ± 0.03 **** 1.45 ± 0.04 0.06 ± 0.03 5.8 ± 0.07
ns
1.52 ± 0.03 0.06 ± 0.05 5.9 ± 0.03 1.69 ± 0.04 0.03 ± 0.02
Lake soil 1.0 ± 0.07*** 1.30 ± 0.02 0.13 ± 0.07 4.2 ± 0.15
ns
1.42 ± 0.04 0.07 ± 0.04 4.4 ± 0.03 1.73 ± 0.04 0.03 ± 0.02
Compost 3.0 ± 0.18 *** 1.44 ± 0.05 0.09 ± 0.06 4.1 ± 0.15 ** 1.50 ± 0.04 0.08 ± 0.04 5.0 ± 0.09 1.74 ± 0.06 0.04 ± 0.05
DNA concentration (μg/mL) was quantified in a spectrophotometer (A
260
nm).
Yield in μg per gram of soil = concentration of DNA (μg/μL). Volume used to suspend DNA (μL) /weight of soil (g).
A
260/280
represents protein contamination. A
340
is an indication for humic acid presence.
PAC represents powdered activated charcoal.
P values were generated by Graph Pad Prism 5.0 software using paired T-test analysis.
P value < 0.05 is considered to be statistically significant. DNA extraction methods performing worse at a statistically significant level as compared with
the best performing method (Glass powder + PAC) are indicated by asterisks
**** indicates P < 0.0001
*** indicates P < 0.001
** indicates P < 0.01
ns
indicates P > 0.05 which is statistically non-significant.
doi:10.1371/journal.pone.0132441.t005
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 10 / 16

Discussion
Various methods are available for metagenom ic DNA extraction based on chemical or
mechanical lysis of microbial cells present in the soil. Among these methods, glass bead beating
is considered to be an effective technique for metagenomic DNA extraction [6]. This method
has also been modified in previous reports to be suitable for different soil types [25]. Commer-
cial kits such as Fast DNA SPIN kit for soil, MP Biomedicals, Santa Ana, CA) and Ultra Clean
Mo Bio Soil DNA isolation kit are also based on the method of bead beating. Even though this
method was adopted for different soils, it requires ‘bead beater’ for cell lysis and the method’s
efficiency depends on the size of beads and duration of agitation. While ‘bead beating’ and our
improved method M6 both utilize glass, the mechanism of cell breakup is fundamentally differ-
ent. Bead mills and other suspension-based cell breakup approaches rely on the fluid-dynamic
shear that resu lts in flows between particles [26], or more accurately, the mechanisms of dissi-
pating the imparted energy at the turbulent length scales of the cells [27]. In contrast, the
improved method M6 utilizes direct mechanical grinding of the soil and glass shards (Table 2).
Therefore, in contra st to shear forces of bead beating, the mortar and pestle grinding represents
a direct physical maceration, which is apparently more effective means of disrupting the cells
in a metagenom ic soil sample based on DNA recovery yield (Table 3). A potential reason for
reduced performance of cell disruption in soil samples by bead beating could result from
increased viscosity due to the presence of a high concentration of insoluble materials during
the beating process [28].
Our improved method M6 involves the fine grinding of soil samples with the glass powder
which takes less than 5 min. In addition to providing an excellent mechanical force for cell
breakup, we suggest that enhanced DNA adsorption on the finely powdered silica may also be
playing a role [18]. Upon grinding, the cells are lysed and the DNA is adsorbed by the silica
particles which are subsequently extracted by addition of extraction buffer [21, 16]. The size of
glass powder varies from 223 to 261 nm. This extremely small size of powdered glass relative to
typical glass milling beads (~100 microns) provides greatly enhanced surface area as well as
freshly fractured silica surfaces to contribute to DNA adsorption. The adsorption of DNA to
silica has been intensely studied in relevance to sequencing analysis and microfluidics [29], and
reports the advantage of large silica surface area for DNA extraction [ 30]. This behavior of
DNA adsorption to glass has the potential to effect the application of these methods for PCR
amplification and sequencing of the environmental samples to study microbial diversity and
functionality in complex communities [16]. Li et al. 2015 [31] reported that the use of silica col-
umns improves viral sequence recovery when compared to other methods of DNA extra ction.
Silica nanoparticles have been fabricated for the effective immobilization and sensitive
sequence-specific detection of DNA [32]. An observation to be validated using the above meth-
odology is its efficiency in the recovery of high molecular weight DNA (>20Kb) for use in
large sized libraries. For this behavior to not affect metagenomic analysis, one must assume
that the process of cell disruption and subsequent shearing of DNA is similar for different
microorganisms. We suggest that the mechanical grinding of the method (M6), as compared to
hydrodynamic shearing described above, may provide for a more representative sampling of
the microbial populations. Future work combining defined mixtures of microorganisms to
sterile soil could address this issue.
The efficiency of using powdered glass in mechanical cell lysis and enhanced surface area is
confirmed by the yield of DNA which is much higher in method M6 than the other methods.
Another advantage of this methodology is the inclusion of PAC in the extraction buffer to min-
imize additional purification steps. PAC is very porous with a vast surface area which allows
the absorption of humic substances , lignin sulphonate, tannic acid, heavy metals and non-
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 11 / 16

degradable coloured substances [33]. PAC has previously been used as a purifying agent in
other reports of soil DNA extraction [11, 22]. In th e present study, 1% PAC was added directly
to the extraction buffer in order to absorb contaminating components of the metagenomic
DNA that would otherwise inhibit subsequent DNA manipulation. This simplifies the need for
subsequent column purification. Polyethylene glycol (30% PEG) has been used in other meta-
genomic DNA extraction metho ds because it does not co-precipitate humic acids with the
DNA as occurs with ethanol or isopropanol precipitation [34]. In the current procedure (M6) ,
the humic substances are precipitated in a simple intermediate step using chloroform and iso-
propanol as reported previously [35].
The powdered glass method (M6) is compared with four other previously reported methods
and a commercial kit. All the methods follow the same basic steps: cell lysis and precipitation
of DNA but they differ from each other on the homogenization approach and duration of the
total protocol (Table 1). Methods M2, M3, M4 and M5 require homogenizing equipment for
cell lysis whereas M1 is based on chemical lysis and M6 is based on glass powder grinding.
Among all the methods studied, powdered glass method (M6) shows higher efficiency of cell
lysis during DNA extraction which is evidenced in terms of DNA yield. The methods were also
tested for four different soil samples that that differed in texture, pH, percentage of organic
matter and the amount of metals (S1 Table). The presence or absence of clay and other particu-
late matter present in each soil is an important consideration because they tend to decrease the
DNA yield by adsorbing the free DNA [36, 37]. Grinding with high surface area powdered
glass is expected to out-compete such adsorption [38]. We feel that this characteristic helps
contribute to the robust performance of method M6 for different soil types.
As noted above, metho ds M1 to M4 did not achieve either high yield or purity. Though
method M1 [39] did not require any sophisticated equipment for cell lysis, it failed to meet
high purity and yield for some of the soil samples, while method M3 apparently suffered from
excessive shearing of the DNA due to sonication. Method M2 based on bead beating is often
recommended as the most effective method; however, in our study, this method did not yield
pure DNA for all the soil samples. Method M2 also requires the bead beater equipment for the
cell lysis. Method M4 that used a combination of lysozyme and freeze thaw did not reduce the
humic acid content. The Fast DNA kit for soil resulted in a better DNA yield and purity but
still contained humic acid residues. Other reports also demonstrate the failure of commercial
resins to provide DNA free from humic acids as they compete with the DNA for column bind-
ing [40 , 41].
Downstream applications such as PCR amplification and restriction digestion involve enzy-
matic reactions that can be significantly inhibited by humic acids. Humic substances chelate
the Mg
2+
ions required for the activity of Taq polymerase, restriction enzymes and ligases [42].
The DNA isolated using methods M1 to M4 did not provide sufficiently pure DNA for further
studies. Kit based method M5 provided DNA suitable for the restriction digestion assay; how-
ever, it was not found suitable for PCR amplification when teste d on different soil samples. The
results reveal that the methods M1 to M5 requires further purification of the DNA for molecu-
lar analysis. The DNA isolated using the powdered glass method M6 displays a high qPCR effi-
ciency; greater than 99%, where the accepted PCR efficiency for qPCR analysis ranges from 90
to 110 percent (Roche Life Sciences, US). The suitability of the DNA for PCR, qPCR, restriction
digestion assay and metagenomic library construction confir ms that sufficient purity was
achieved using method M6; a simplified method that integrates cell lysis and DNA purification
steps (Table 2).
Despite the diversity in the sources of the soil samples, our improve d powdered glass
method (M6) produced high yield of DNA, demonstrating broader applicability of the method.
The average DNA yield of the four soil samples was 5.87 μg/g which was considerably higher
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 12 / 16

than the other methods (Table 1). In addition, protein and humic acid contamination was low
when compared to the other tested methods. The purity of the metagenomic DNA obtained by
this method M6 was further substantiated by facile construction of a metagenomic library. Fur-
ther, this method was also successfully used for DNA extraction from Gram positive, Gram
negative bacteria and also microalgae. The statistically significant improved efficiency of DNA
extraction invariably contributes to downstream manipulations by effectively reducing the
contaminants.
As a final added advantage, the overall processing time for powdered glass method (M6)
was shorter than the other tested methods. The earlier reported methods (M1 to M4) took
about 5 to 7 h for proc essing while the method M6 requires only 1.5 h which was considerably
shorter than the recently reported method of Sagar et al. (2014) [35]. The commercial kit
(method M5) took the least time for DNA extraction (Table 1); however, the cost of processing
1 g of soil for a single reaction is about $8 (US) which is quite high if large numbers of samples
are to be processed. The use of glass powder is an economical method for cell lysis that does
not require sophisticated equipment or enzymes for cell lysis.
Conclusion
A robust method of DNA extraction from soil for metagenomic analysis and cultivated organ-
isms is demonstrated which uses glass powder for cell lysis and powdered activated carbon for
purification. This method is rapid, efficient and economical, and found to be robustly applica-
ble for DNA extraction where intact and pure DNA is requir ed, and may include additional
advantages for providing a representative sampling of the metabiome population.
Supporting Information
S1 Fig. Size determination of the glass powder by FE-SEM analysis.
(DOC)
S2 Fig. Gel electrophoresis of genomic DNA isolated by method M6 for Gram positive,
Gram negative bacteria and microalgae .
(DOC)
S3 Fig. Gel electrophoresis of PCR amplified 16S rDNA for the metagenomic DNA
extracted by methods M1 to M5.
(DOC)
S4 Fig. Gel electrophoresis of partial restriction digestion of the metagenomic DNA
extracted by methods M1, M2, M4 and M5 usi ng BamHI.
(DOC)
S5 Fig. Gel electrophoresis of metagenomic DNA extraction by the glass beads, glass pow-
der and glass powder + powdered activated charcoal for four different soils.
(DOC)
S6 Fig. qPCR analysis for method M6.
(DOC)
S1 Table. Characteristics of the four different soil samples.
(DOC)
S2 Table. Suitabi lity of the DNA samples extracted by methods M1 to M6 for 16S rDNA
PCR amplification and partial restriction digestion by BamHI.
(DOC)
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 13 / 16

S3 Table. Triplicate values for DNA DNA yield (μg/g of soil) for methods M1 to M6.
(DOC)
S4 Table. Triplicate values for A
260/280
.
(DOC)
S5 Table. Triplicate values for A
340
.
(DOC)
S6 Table. Triplicate values for DNA yield (μg/g of soil) for method M2 and its modifica-
tion.
(DOC)
Acknowledgments
The authors acknowledge SRM University for the infrastructure facilities.
Author Contributions
Conceived and designed the experiments: MR. Performed the experiments: SGD SR AAF RA.
Analyzed the data: SGD MR. Contributed reagents/materials/analysis tools: SR AAF. Wrote
the paper: WRC SGD.
References
1. Burgmann H, Pesaro M, Widmer F, Zeyer J. A strategy for optimizing quality and quantity of DNA
extracted from soil. J Microbiol Meth. 2001; 45: 7–20.
2. Cottrell MT, Waidner L, Yu L, Kirchman DL. Bacterial diversity of metagenomic and PCR libraries from
the Delaware River. Environ Microbiol. 2005; 7: 1883–1895. PMID: 16309387
3. Gray JP, Herwig RP. Phylogenetic analysis of the bacterial communities in marine sediments. Appl
Environ Microbiol. 1996; 62: 4049–4059. PMID: 8899989
4. Jacobsen CS, Rasmussen OF. Development and application of a new method to extract bacterial DNA
from soil based on separation of bacteria from soil with cation-exchange resin. Appl Environ Microbiol.
1992; 58: 2458–2462. PMID: 16348750
5. Kauffmann IM, Schmitt J, Schmid RD. DNA isolation from soil samples for cloning in different hosts.
Appl Microbiol Biotechnol. 2004; 64: 665–670. PMID: 14758515
6. Yeates C, Gillings MR, Davison D, Altavilla N, Veal D. Methods for microbial DNA extraction from soil
for PCR amplification. Biol Proced Online. 1998; 1: 40–47. PMID: 12734590
7. Purohit MK, Singh SP. Assessment of various methods for extraction of metagenomic DNA from saline
habitats of coastal Gujarat (India) to explore molecular diversity. Lett Appl Microbiol. 2009; 49: 338–
344. doi: 10.1111/j.1472-765X.2009.02663.x PMID: 19712192
8. Johnston CG, Aust SD. Detection of Phanerochaetechrysosporium in soil by PCR and restriction
enzyme analysis. Appl Environ Microbiol. 1994; 60: 2350–2354. PMID: 8074515
9. Lee S, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial
strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996; 62: 3787–3793. PMID:
8837435
10. Tsai Y, Olson BH. Rapid method for direct extraction of DNA from soil and sediment.Appl Environ
Microbiol. 1991; 57:1070–1074. PMID: 1647749
11. Verma D, Satyanarayana T. An improved protocol for DNA extraction from alkaline soil and sediment
samples for constructing metagenomic libraries. Appl Biochem Biotechnol. 2011; 165: 454–464. doi:
10.1007/s12010-011-9264-5 PMID: 21519906
12. Wise MG, McArthur IY, Shimkets L. Bacterial diversity of a Carolina Bay as determined by 16S rRNA
gene analysis: confirmation of novel taxa. Appl Environ Microbiol. 1997; 63: 1505–1514. PMID:
9097448
13. Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and puri-
fication procedures for soil and sediment samples Appl Environ Microbiol. 1999; 65: 4715–4724.
PMID: 10543776
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 14 / 16

14. Sharma PK, Capalash N, Kaur J. An improved method for single step purification of metagenomic
DNA. Mol Biotechnol. 2007; 36: 61–63. PMID: 17827539
15. Liesack W, Stackebrandt E. Occurrence of novel groups of the domain bacteria as revealed by analysis
of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992; 174: 5072–
5078. PMID: 1629164
16. Rojas-Herrera R, Narvaez-Zapata J, Zamudio-Maya M, Mena-Martinez ME. A simple silica-based
method for metagenomic DNA extraction from soil and sediments. Mol Biotechnol. 2008; 40: 13–17.
doi: 10.1007/s12033-008-9061-8 PMID: 18373226
17. Attal J, Cajero-Juarez M, Louis-Marie H. A simple method of DNA extraction from whole tissues and
blood using glass powder for detection of transgenic animals by PCR. Transgenic Res. 1995; 4: 149–
150. PMID: 7704054
18. Marko MA, Chipperfield R, Birnboim HC. A procedure for the large-scale isolation of highly purified plas-
mid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982; 121: 382–387.
PMID: 6179438
19. Punekar NS, Suresh Kumar SV, Jayashri TN, Anuradha R. Isolation of genomic DNA from acetone-
dried Aspergillus mycelia. Fungal Genet News. 2003; 50:15–16.
20. Hussain T, Singh BP, Anwar F. A quantitative Real Time PCR based method for the detection of Phy-
tophthora infestans causing Late blight of potato, in infested soil. Saudi J Biol Sci. 2014; 21: 380–386.
doi: 10.1016/j.sjbs.2013.09.012 PMID: 25183949
21. Radha S, Aliya Fathima A, Iyappan S, Ramya M. Direct colony PCR for rapid identification of varied
microalgae from freshwater environment. J Appl Phycol. 2013; 25: 609–613.
22. Sharma S, Sharma KK, Kuhad RC. An efficient and economical method for extraction of DNA amena-
ble to biotechnological manipulations, from diverse soils and sediments. J Appl Microbiol. 2014; 116:
923–933. doi: 10.1111/jam.12420 PMID: 24329912
23. APHA, AWWA, WEF. Standard methods for the examination of water and wastewater. 21sted. Wash-
ington DC: American Public Health Association; 2005.
24. Nair HP, Vincent H, Bhat SG. Evaluation of five in situ lysis protocols for PCR amenable metagenomic
DNA from mangrove soils. Biotechnol Rep. 2014; 4: 134–138.
25. Siddhapura PK, Vanparia S, Purohit MK, Singh SP. Comparative studies on the extraction of metage-
nomic DNA from the saline habitats of coastal Gujarat and Sambhar Lake, Rajasthan (India) in prospect
of molecular diversity and search for novel biocatalysts. Int J Biol Macromol. 2010; 47: 375–379. doi:
10.1016/j.ijbiomac.2010.06.004 PMID: 20600268
26. Doebler RW, Erwin B, Hickerson A, Irvine B, Woyski D, Nadim A, et al. Continuous-flow, rapid lysis
devices for biodefense nucleic acid diagnostic systems. J Assoc Lab Autom. 2009; 14:119–125.
27. Aziz A, Werner BC, Epting KL, Agosti CD, Curtis WR. The cumulative and sub-lethal effects of turbu-
lence on erythrocytes in a stirred-tank model. Ann Biomed Eng. 2007; 35: 2108–2120. Available: http://
engineering.http://engineering. PMID: 17909969
28. Curtis WR, Emery AH. Plant cell suspension culture rheology. Biotechnol Bioengg. 1993; 42: 520–526.
29. Poeckh T, Lopez S, Fuller AO, Solomon MJ, Larson RG. Adsorption and elution characteristics of
nucleic acids on silica surfaces and their use in designing a miniaturized purification unit. Anal Biochem.
2008; 373: 253–262. Available: http://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=
pubmed&retmode=ref&cmd=prlinks&id=18022378. PMID: 18022378
30. Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and
simple method for purification of nucleic acids. J Clin Microbiol. 1990; 28: 495–503. PMID: 1691208
31. Li L, Deng X, Mee ET, Collot-Teixeira S, Anderson R, Schepelmann S, et al. Comparing viral metage-
nomics methods using a highly multiplexed human viral pathogens reagent. J Virol Methods. 2015;
213: 139–146. doi: 10.1016/j.jviromet.2014.12.002 PMID: 25497414
32. Zhang D, Chen Y, Chen HY, Xia XH. Silica-nanoparticle-based interface for the enhanced immobiliza-
tion and sequence-specific detection of DNA. Anal Bioanal Chem. 2004 379: 1025–1030. PMID:
15179534
33. Seo GT, Ohgaki S. Evaluation of refractory organic removal in combined biological powdered activated
carbon-microfiltration for advanced wastewater treatment. Water Sci Technol. 2001; 43: 67–74.
34. La Montagne MG, Michel FC, Holden PA, Reddy CA. Evaluation of extraction and purification methods
for obtaining PCR-amplifiable DNA from compost for microbial community analysis. J Microbiol Meth.
2002; 49: 255–264.
35. Sagar K, Singh SP, Goutam KK, Konwar BK. Assessment of five soil DNA extraction methods and a
rapid laboratory-developed method for quality soil DNA extraction for 16S rDNA-based amplification
and library construction. J Microbiol Meth. 2014; 97: 68–73.
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 15 / 16

36. Frostegard A, Courtois V, Ramisse S, Clerc D, Bernillon F, Le GP, et al. Quantification of bias related to
the extraction of DNA directly from soils. Appl Environ Microbiol. 1999; 65: 5409–5420. PMID:
10583997
37. Roose-Amsaleg CL, Garnier SE, Harry M. Extraction and purification of microbial DNA from soil and
sediment samples. Appl Soil Ecol. 2001; 18: 47–60.
38. Buscot F. Springer-Verlag. In: Buscot F, Varma A, editors. Microorganisms in soils: roles in genesis
and functions. Heidelberg; 2005. pp. 19–58.
39. Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol.
1996; 62: 316–322. PMID: 8593035
40. Harry M, Gambier B, Bourezgui Y, Garnier SE. Evaluation of purification procedures for DNA extracted
from organic rich samples. Interference with humic substances. Analusis. 1999; 27: 439–442.
41. Tiedje JM, Asuming-Brempong S, NuÈsslein K, Marsh TL, Flynn SJ. Opening the black box of soil
microbial diversity. Appl Soil Ecol. 1999; 13: 109–122.
42. Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic
resistance genes. Environ Microbiol. 2004; 6: 981–989. PMID: 15305923
Metagenomic DNA Extraction
PLOS ONE | DOI:10.1371/journal.pone.0132441 July 13, 2015 16 / 16
